Basic Usage
This tutorial will demonstrate the most basic Gryffin functionality. We will optimize a single parameter with respect to a simple one dimensional objective function.
[1]:
import os
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
import numpy as np
import pandas as pd
from gryffin import Gryffin
import matplotlib.pyplot as plt
import matplotlib
%matplotlib inline
import seaborn as sns
sns.set(context='talk', style='ticks')
ERROR:root:No traceback has been produced, nothing to debug.
Let’s define the objective function and a helper function that will parse the Gryffin param dict for us:
[2]:
def objective(x):
def sigmoid(x, l, k, x0):
return l / (1 + np.exp(-k*(x-x0)))
sigs = [sigmoid(x, -1, 40, 0.2),
sigmoid(x, 1, 40, 0.4),
sigmoid(x, -0.7, 50, 0.6),
sigmoid(x, 0.7, 50, 0.9)
]
return np.sum(sigs, axis=0) + 1
def compute_objective(param):
x = param['x']
param['obj'] = objective(x)
return param
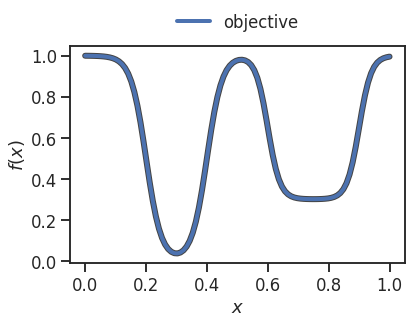
Unlike in most use-cases, we can now visualize the objective function:
[3]:
x = np.linspace(0, 1, 100)
_ = plt.plot(x, objective(x), linewidth=6, color='#444444')
_ = plt.plot(x, objective(x), linewidth=4, label='objective')
_ = plt.xlabel('$x$')
_ = plt.ylabel('$f(x)$')
_ = plt.legend(loc='lower center', ncol=2, bbox_to_anchor=(0.5 ,1.), frameon=False)
We define Gryffin’s configuration as a dictionary. Note that the config can also be passed into Gryffin as a json file using the config_file argument. A continuous parameter x is defined and bounded by 0.0 < x < 1.0. We also define a minimization objective.
[4]:
config = {
"parameters": [
{"name": "x", "type": "continuous", "low": 0., "high": 1., "size": 1}
],
"objectives": [
{"name": "obj", "goal": "min"}
]
}
Gryffin’s instance can now be initialized with the configuration above. Here we select silent=True to suppress the rich display in the notebook. Only warnings and errors will be printed.
[5]:
gryffin = Gryffin(config_dict=config, silent=True)
In the cell below, we perform a sequential optimization for a maximum of 15 evaluations.
[6]:
%debug
observations = []
MAX_ITER = 15
for num_iter in range(MAX_ITER):
print('-'*20, 'Iteration:', num_iter+1, '-'*20)
# Query for new parameters
params = gryffin.recommend(observations=observations)
# Params is a list of dict, where each dict containts the proposed parameter values, e.g., {'x':0.5}
# in this example, len(params) == 1 and we select the single set of parameters proposed
param = params[0]
print(' Proposed Parameters:', param, end=' ')
# Evaluate the proposed parameters. "compute_objective" takes param, which is a dict, and adds the key "obj" with the
# objective function value
observation = compute_objective(param)
print('==> :', observation['obj'])
# Append this observation to the previous experiments
observations.append(observation)
-------------------- Iteration: 1 --------------------
Could not find any observations, falling back to random sampling
Proposed Parameters: {'x': 0.85985327} ==> : 0.3829059252850222
-------------------- Iteration: 2 --------------------
{'bias_loc': 0.0, 'bias_scale': 1.0, 'hidden_shape': 6, 'learning_rate': 0.05, 'num_draws': 1000, 'num_epochs': 2000, 'num_layers': 3, 'weight_loc': 0.0, 'weight_scale': 1.0}
here
Sequential(
(linear1): BayesLinear(prior_mu=0.0, prior_sigma=1.0, in_features=1, out_features=6, bias=True)
(relu1): ReLU()
(linear2): BayesLinear(prior_mu=0.0, prior_sigma=1.0, in_features=6, out_features=1, bias=True)
)
BayesLinear(prior_mu=0.0, prior_sigma=1.0, in_features=1, out_features=6, bias=True)
here2
ReLU()
BayesLinear(prior_mu=0.0, prior_sigma=1.0, in_features=6, out_features=1, bias=True)
here2
{'weight_0': tensor([[-1.6142, -0.8541, -1.0287, -0.8812, -0.0362, -0.9161],
[-1.6296, -2.0786, -0.9523, -0.8295, -0.2203, -1.6839],
[-1.4362, -0.8384, -1.4848, -0.8497, -0.8624, -1.0217],
...,
[-1.7061, -1.6743, -0.9764, -1.0882, -1.4251, -1.0485],
[-1.6801, -1.4916, -0.9102, -0.7150, -1.0392, -1.2135],
[-1.5762, -1.2191, -1.5172, -1.0660, -1.1869, -0.7096]]), 'bias_0': tensor([[-1.5404, -0.8772, -1.0658, -0.6388, -0.9345, -0.7542],
[-1.8432, -1.4672, -1.0981, -0.7036, -0.7027, -0.9699],
[-1.5911, -1.0813, -1.4145, -0.8012, -0.9155, -0.6964],
...,
[-1.6194, -1.0552, -0.7723, -0.6375, -0.8930, -0.7536],
[-1.6779, -1.3234, -1.1714, -0.7849, -1.1290, -0.6910],
[-1.7789, -1.3437, -0.9787, -0.9873, -0.7164, -0.8679]]), 'weight_1': tensor([[ 0.5331, 1.7729, 1.1227, 1.1296, 0.9411, 0.6728],
[ 0.8856, 0.1455, 0.6103, -0.7441, 0.6350, -0.0995],
[ 0.2199, -0.1605, 0.1302, 0.3524, -0.8872, 0.8343],
...,
[ 0.3596, -0.3079, -0.0453, -0.5165, 0.1554, 0.4110],
[ 0.8396, -0.2201, -0.7451, 1.6703, -0.3121, -0.1851],
[ 0.7888, 0.4437, -0.3611, 0.6328, 1.2197, 1.0933]]), 'bias_1': tensor([[ 0.5223, 0.4054, 0.0613, 0.0558, 0.0763, 0.1591],
[ 0.3594, 0.5762, 0.3886, -0.0126, 0.1548, 0.1460],
[ 0.8739, -0.0381, 0.6748, -0.0945, -0.0758, 0.5327],
...,
[ 0.5928, 0.5441, 0.8780, -0.0446, -0.0041, 0.4008],
[ 0.7220, 0.8011, -0.0934, 0.0332, -0.1782, 0.3133],
[ 1.0521, 0.2524, 0.4532, -0.0044, 0.3840, 0.5770]])}
here3
dict_keys(['weight_0', 'bias_0', 'weight_1', 'bias_1', 'gamma'])
0
tensor([[-1.6142, -0.8541, -1.0287, -0.8812, -0.0362, -0.9161],
[-1.6296, -2.0786, -0.9523, -0.8295, -0.2203, -1.6839],
[-1.4362, -0.8384, -1.4848, -0.8497, -0.8624, -1.0217],
...,
[-1.7061, -1.6743, -0.9764, -1.0882, -1.4251, -1.0485],
[-1.6801, -1.4916, -0.9102, -0.7150, -1.0392, -1.2135],
[-1.5762, -1.2191, -1.5172, -1.0660, -1.1869, -0.7096]])
tensor([[0.8599]])
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Input In [6], in <cell line: 4>()
5 print('-'*20, 'Iteration:', num_iter+1, '-'*20)
7 # Query for new parameters
----> 8 params = gryffin.recommend(observations=observations)
10 # Params is a list of dict, where each dict containts the proposed parameter values, e.g., {'x':0.5}
11 # in this example, len(params) == 1 and we select the single set of parameters proposed
12 param = params[0]
File /ssd003/home/jwilles/gryffin/src/gryffin/gryffin.py:318, in Gryffin.recommend(self, observations, sampling_strategies, num_batches, as_array)
314 # ----------------------------------------------
315 # sample bnn to get kernels for all observations
316 # ----------------------------------------------
317 self.log_chapter('Bayesian Network')
--> 318 self.bayesian_network.sample(obs_params) # infer kernel densities
319 # build kernel smoothing/classification surrogates
320 self.bayesian_network.build_kernels(descriptors_kwn=descriptors_kwn, descriptors_feas=descriptors_feas,
321 obs_objs=obs_objs, obs_feas=obs_feas, mask_kwn=mask_kwn)
File /ssd003/home/jwilles/gryffin/src/gryffin/bayesian_network/bayesian_network.py:148, in BayesianNetwork.sample(self, obs_params)
140 model = trainer.train(obs_params)
141 # trace_kernels = run_tf_network(
142 # observed_params=obs_params,
143 # frac_feas=self.frac_feas,
144 # config=self.config,
145 # model_details=self.model_details,
146 # )
--> 148 self.trace_kernels = model.get_kernels()
151 # self.trace_kernels = trace_kernels
153 end = time.time()
File /ssd003/home/jwilles/gryffin/src/gryffin/bayesian_network/torch_interface/bnn.py:216, in BNN.get_kernels(self, num_draws)
214 if num_draws == None:
215 num_draws = self.num_draws
--> 216 self._sample(num_draws)
218 print(self.trace)
220 trace_kernels = {'locs': [], 'sqrt_precs': [], 'probs': []}
File /ssd003/home/jwilles/gryffin/src/gryffin/bayesian_network/torch_interface/bnn.py:203, in BNN._sample(self, num_draws)
201 print('here3')
202 print(posterior_samples.keys())
--> 203 post_kernels = self.numpy_graph.compute_kernels(posterior_samples, self.frac_feas)
205 self.trace = {}
206 for key in post_kernels.keys():
File /ssd003/home/jwilles/gryffin/src/gryffin/bayesian_network/tfprob_interface/numpy_graph.py:56, in NumpyGraph.compute_kernels(self, posteriors, frac_feas)
54 single_bias = bias[sample_index]
55 print(post_layer_outputs[-1][sample_index])
---> 56 output = activation( np.matmul( post_layer_outputs[-1][sample_index], single_weight) + single_bias)
57 outputs.append(output)
59 post_layer_output = np.array(outputs)
ValueError: matmul: Input operand 1 has a mismatch in its core dimension 0, with gufunc signature (n?,k),(k,m?)->(n?,m?) (size 6 is different from 1)
Let’s now plot all the parameters that have been probed and the best solution found.
[ ]:
# objective function
x = np.linspace(0, 1, 100)
y = [objective(x_i) for x_i in x]
# observed parameters and objectives
samples_x = [obs['x'] for obs in observations]
samples_y = [obs['obj'] for obs in observations]
_ = plt.plot(x, y, linewidth=6, color='#444444')
_ = plt.plot(x, y, linewidth=4, label='objective')
_ = plt.scatter(samples_x, samples_y, zorder=10, s=150, color='r', edgecolor='#444444', label='samples')
# highlight best
_ = plt.scatter(samples_x[np.argmin(samples_y)], np.min(samples_y), zorder=11, s=150,
color='yellow', edgecolor='#444444', label='best')
# labels
_ = plt.xlabel('$x$')
_ = plt.ylabel('$f(x)$')
_ = plt.legend(loc='lower center', ncol=3, bbox_to_anchor=(0.5 ,1.), frameon=False)
[ ]: