Perovskites
In this tutorial, we will introduce optimization of categorical parameters using Gryffin using a real-world application from materials science. Namely, we will consider optimization of the bandgap of hybrid organic-inorganic perovskite (HOIP) solar cells. This example is taken directly from Section V.B. in the Gryffin paper entitled Discovery of hybrid oraginc-inorganic perovskies
Perovskites solar cells are a promising class of ligh harvesting materials and are typically comprised of inorganic lead halide matrices and contain inorganic or organic anions. This application concerns the design of of hybrid organic-inorganic perovskites (HOIPs) based on a recently reported dataset. The HOIP candidates of this dataset are designed from a set of four different halide anions, three different group-IV cations and 16 different organic anions, resulting in 192 unique HOIP candidate materials.
This example will also detail the use of physicochemical descriptors for the options of categorical variables to further accelerate the optimization rate of Gryffin.
[1]:
import pickle
import numpy as np
import pandas as pd
import os
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
[2]:
from gryffin import Gryffin
First, we will set some variables that will parameterize our optimization campaign. budget indicates the number of bandgap measurements we intend on making. sampling_strategies represent the values of \(\lambda\) used by Gryffin’s acquisition function. with_desc indicates whether or not to utilize the descriptors of the perovskite components. dynamic indicates whether or not to use the dynamic formulation of Gryffin to refine the provided descriptors.
[3]:
budget = 192
sampling_strategies = np.array([-1, 1])
with_desc = True
dynamic = True
random_seed = 2020
[4]:
# the categorical options corresponding to the minimum bandgap in the dataset (optimum)
optimum = ['hydrazinium', 'I', 'Sn'] # value = 1.5249 eV
[5]:
# load in the perovskites dataset as a pandas DataFrame
lookup_df = pickle.load(open('perovskites.pkl', 'rb'))
[6]:
# helper functions
def measure(param):
''' lookup the HSEO6 bandgap for given perovskite composition
'''
match = lookup_df.loc[
(lookup_df.organic == param['organic']) &
(lookup_df.anion == param['anion']) &
(lookup_df.cation == param['cation'])
]
assert len(match)==1
bandgap = match.loc[:, 'hse06'].to_numpy()[0]
return bandgap
def get_descriptors(element, kind):
''' retrive the descriptors for a given categorical variable option
'''
return lookup_df.loc[(lookup_df[kind]==element)].loc[:, lookup_df.columns.str.startswith(f'{kind}-')].values[0].tolist()
We will now prepare the descriptors for use with Gryffin. The three categorical variables which together comprise a perovskite material are
halide anions,
anion(4 options)group-IV cations,
cation(3 options)organic anions,
organic(16 options)
A depiction of the HOIP space is shown below.
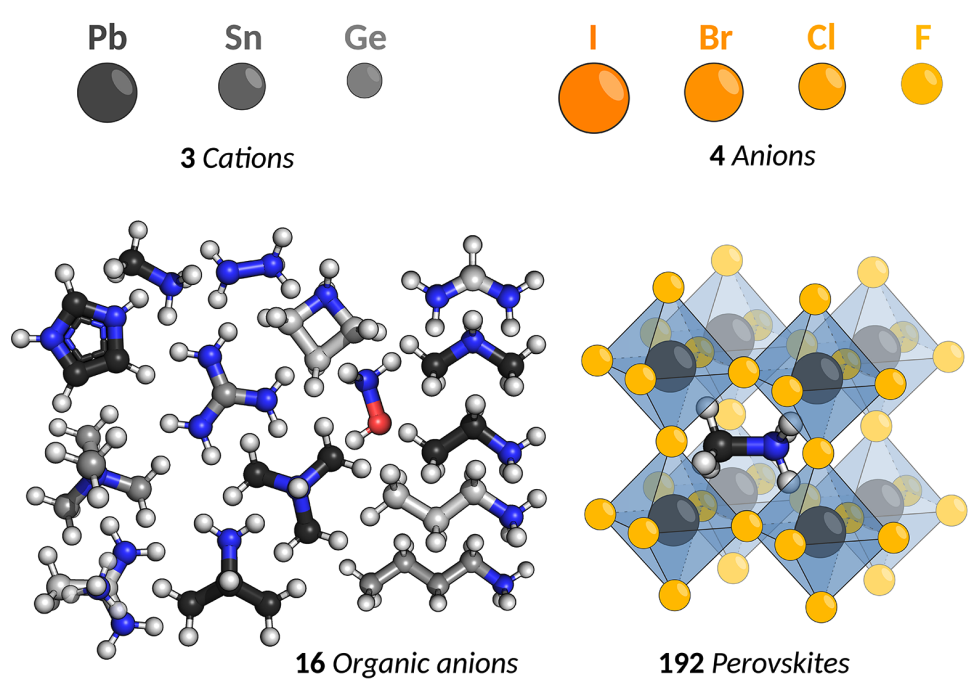
We characterize the inorganic constituents (anion and cation) by their electron affinity, ionization energy, mass, and electronegatvity. Organic components are described by their HOMO and LUMO energies, dipole moment, atomization energy, radius of gyration, and molecular weight.
For categorcial variables with descriptors, Gryffin accepts a dictonary of descriptors in the following form
descriptors = {'option_0_name': [option_0_desc_0, option_0_desc_1, ...], 'option_1_name':, [...], ...}
For the naive formulation of Gryffin (essentially one-hot-encoding of categorical variables), one should use the following descriptor format
descriptors = {'option_0_name': None, 'option_1_name':, None, ...}
The dictionary descritptors can then be passed into the Gryffin config as shown below (with the key category_details)
[7]:
# prepare descriptors
organic_options = lookup_df.organic.unique().tolist()
anion_options = lookup_df.anion.unique().tolist()
cation_options = lookup_df.cation.unique().tolist()
[8]:
if with_desc:
# use physicochemical descriptors - static or dynamic gryffin
desc_organic = {option: get_descriptors(option, 'organic') for option in organic_options}
desc_anion = {option: get_descriptors(option, 'anion') for option in anion_options}
desc_cation = {option: get_descriptors(option, 'cation') for option in cation_options}
else:
# no descriptors - naive gryffin
desc_organic = {option: None for option in organic_options}
desc_anion = {option: None for option in anion_options}
desc_cation = {option: None for option in cation_options}
[9]:
# gryffin config
config = {
"general": {
"num_cpus": 4,
"auto_desc_gen": dynamic,
"batches": 1,
"sampling_strategies": 1,
"boosted": False,
"caching": True,
"random_seed": random_seed,
"acquisition_optimizer": "genetic",
"verbosity": 3
},
"parameters": [
{"name": "organic", "type": "categorical", 'options': organic_options, 'category_details': desc_organic},
{"name": "anion", "type": "categorical", 'options': anion_options, 'category_details': desc_anion},
{"name": "cation", "type": "categorical", 'options': cation_options, 'category_details': desc_cation},
],
"objectives": [
{"name": "bandgap", "goal": "min"},
]
}
Once we have set the config, we are ready to commence with the optimization campaign. Here, we measure perovskite bandgaps sequentially (one-at-a-time) using alternating sampling strategies which in this case corresponds to alternating exploitative/explorative behaviour. We continue the optimization until we reach the global optimum (defined above) or we exhaust our budget.
[11]:
observations = []
# initialize gryffin
gryffin = Gryffin(config_dict=config, silent=True)
for num_iter in range(budget):
print('-'*20, 'Iteration:', num_iter+1, '-'*20)
# alternating sampling strategies, assuming batch size of 1
idx = num_iter % len(sampling_strategies)
sampling_strategy = sampling_strategies[idx]
# ask Gryffin for a new sample
samples = gryffin.recommend(observations=observations, sampling_strategies=[sampling_strategy])
measurement = measure(samples[0])
samples[0]['bandgap'] = measurement
observations.extend(samples)
print(f'SAMPLES : {samples}')
print(f'MEASUREMENT : {measurement}')
# print(f'ITER : {num_iter}\tSAMPLES : {samples}\t MEASUREMENT : {measurement}')
# check for convergence
if [samples[0]['organic'], samples[0]['anion'], samples[0]['cation']] == optimum:
print(f'FOUND OPTIMUM AFTER {num_iter+1} ITERATIONS!')
break
-------------------- Iteration: 1 --------------------
Could not find any observations, falling back to random sampling
SAMPLES : [{'organic': 'dimethylammonium', 'anion': 'Cl', 'cation': 'Pb', 'bandgap': 3.3139}]
MEASUREMENT : 3.3139
-------------------- Iteration: 2 --------------------
SAMPLES : [{'organic': 'ethylammonium', 'anion': 'Br', 'cation': 'Pb', 'bandgap': 2.5996}]
MEASUREMENT : 2.5996
-------------------- Iteration: 3 --------------------
SAMPLES : [{'organic': 'ammonium', 'anion': 'F', 'cation': 'Sn', 'bandgap': 4.9081}]
MEASUREMENT : 4.9081
-------------------- Iteration: 4 --------------------
SAMPLES : [{'organic': 'ethylammonium', 'anion': 'I', 'cation': 'Pb', 'bandgap': 2.2316}]
MEASUREMENT : 2.2316
-------------------- Iteration: 5 --------------------
SAMPLES : [{'organic': 'butylammonium', 'anion': 'F', 'cation': 'Sn', 'bandgap': 4.8539}]
MEASUREMENT : 4.8539
-------------------- Iteration: 6 --------------------
SAMPLES : [{'organic': 'guanidinium', 'anion': 'I', 'cation': 'Pb', 'bandgap': 2.3586}]
MEASUREMENT : 2.3586
-------------------- Iteration: 7 --------------------
SAMPLES : [{'organic': 'tetramethylammonium', 'anion': 'I', 'cation': 'Pb', 'bandgap': 2.5415}]
MEASUREMENT : 2.5415
-------------------- Iteration: 8 --------------------
SAMPLES : [{'organic': 'propylammonium', 'anion': 'I', 'cation': 'Pb', 'bandgap': 2.52}]
MEASUREMENT : 2.52
-------------------- Iteration: 9 --------------------
SAMPLES : [{'organic': 'hydrazinium', 'anion': 'I', 'cation': 'Ge', 'bandgap': 1.8829}]
MEASUREMENT : 1.8829
-------------------- Iteration: 10 --------------------
SAMPLES : [{'organic': 'hydrazinium', 'anion': 'Br', 'cation': 'Pb', 'bandgap': 3.1859}]
MEASUREMENT : 3.1859
-------------------- Iteration: 11 --------------------
SAMPLES : [{'organic': 'hydrazinium', 'anion': 'I', 'cation': 'Sn', 'bandgap': 1.5249}]
MEASUREMENT : 1.5249
FOUND OPTIMUM AFTER 11 ITERATIONS!
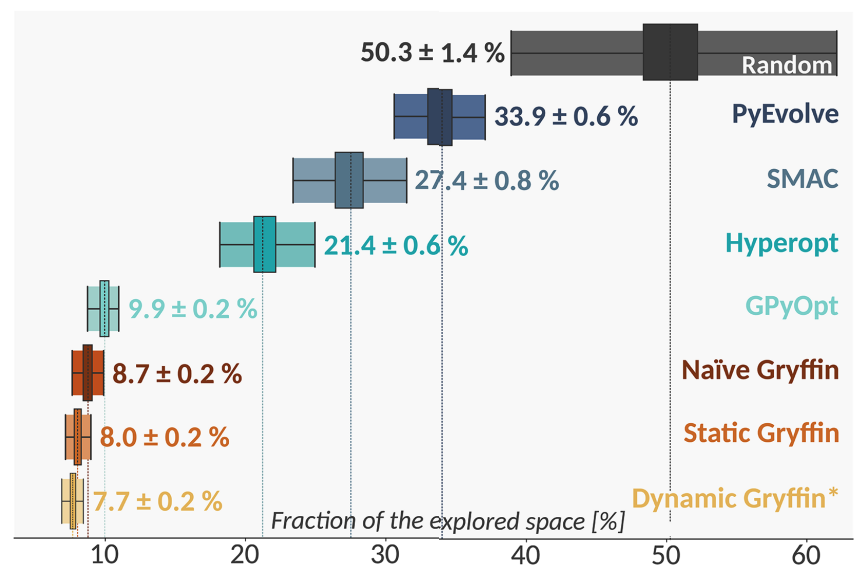
After repeated executions of this experiment, one can compare the average performance of different optimization strategies based on their ability to efficeintly identify HOIP candidates with promising bandgaps. For instance, in the plot below we compare various optimization strategies using the percentage of the parameter space explored before identifying the candidate with the smallest bandgap. Efficient optimizers will thus need to explore a smaller fraction of the space before measuring the optimum.